Describe an application of cloud computing technology in the field of biology.
BIOLOGY: PROTEIN STRUCTURE PREDICTION (PSP)
- Cloud computing is a new technology that allows users to access a variety of computer services on demand. It gives users easy access to a pool of higher-level services and other system resources. Cloud computing has become more important in the realms of geology, biology, and other scientific studies.
- The finest example of a study area that uses cloud technologies for processing and storage is protein structure prediction. A protein is made up of peptide bonds that connect lengthy sequences of amino acids. The varied structures of proteins aid in the development of novel therapeutics, and Protein structure prediction is the prediction of various sequences of proteins based on their three-dimensional structure.
- Protein primary structures are created first, and secondary, tertiary, and quaternary structures are predicted from the fundamental structure. Protein structural predictions are made in this manner. Protein structure prediction employs a variety of different technologies, including artificial neural networks, artificial intelligence, machine learning, and probabilistic approaches, and is crucial in disciplines such as theoretical chemistry and bioinformatics.
Protein
- Proteins are long chains of amino acids that form the basis of all life. They are large molecules that our cells need to function properly. They consist of amino acids. The structure and function of our bodies depend on proteins. The regulation of the body's cells, tissues, and organs cannot happen without them.
- Muscles, skin, bones, and other parts of the human body contain significant amounts of protein, including enzymes, hormones, and antibodies.
- The human body consists of around 100 trillion cells. Each cell has thousands of different proteins. Together, these cause each cell to do its job. The proteins are like tiny machines inside the cell.
Why cloud computing for PSP?
- It requires high computing capabilities and often operates on large data- sets that cause extensive I/O operations.
- Protein structure prediction is a computationally intensive task that is fundamental to different types of research in the life sciences.
- Manually 3D structure determination is difficult, slow, and expensive.
- Structure helps in the design of new drugs for the treatment of diseases.
- The geometric structure of a protein cannot be directly inferred from the sequence of genes that compose its structure, but it is the result of complex computations aimed at identifying the structure that minimizes the required energy. While doing so, high computational power is required which is extremely expensive to own.
- Cloud computing grants access to such capacity on pay per use basis.
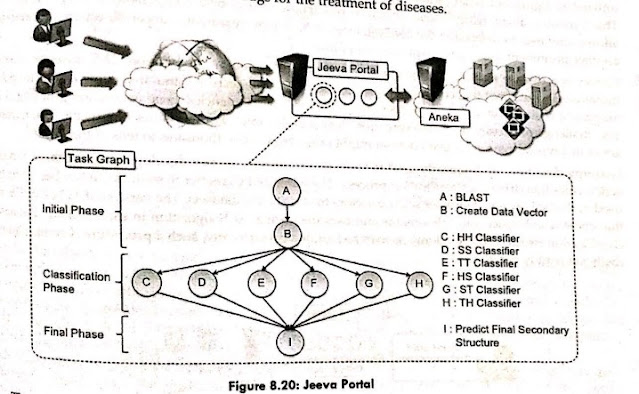
- Jeeva, an integrated Web site that allows scientists to outsource the prediction process to a computing cloud based on Aneka, is one project that studies the use of cloud technology for protein structure prediction. Machine learning approaches are used in the prediction job to determine the secondary structure of proteins. These methods turn the problem into a pattern recognition issue, in which a sequence must be sorted into one of three categories (E, H, and C). The pattern recognition issue is divided into three steps by a popular method based on support vector machines: initialization, classification, and a final phase.
- Even though all three stages must be completed in order, it is possible to use parallel execution in the classification step, where many classifiers are run simultaneously. This opens up the possibility of reducing the prediction's computing time sensibly. After that, the prediction method is converted into a task graph, which is then sent to Aneka. The middleware makes the findings accessible for display through the portal once the task is done.
- Protein structure prediction may be done using a variety of techniques and methods. CASP (Critical Assessment of Protein Structure Prediction) is a well-known technology that gives ways for automated web servers, and research findings are stored in clouds such as the CAMEO (Continuous Automated Model Evaluation) server. These servers may be accessible by anybody at any time and from any location, depending on their needs. Phobius, FoldX, LOMETS, Prime, Predict Protein, SignalP, BBSP, EVfold, Biskit, HHpred, Phone, and ESyired 3D are some of the tools or services used in protein structure prediction. New structures are anticipated using these technologies, and the findings are stored on cloud-based. servers.
Biology: Gene Expression Data Analysis
- Gene expression profiling is the simultaneous assessment of thousands of genes' expression levels. It is utilized to figure out what biological processes are activated at the cellular level by medicinal therapy. This function, along with protein structure prediction, is a critical component of drug design since it allows scientists to determine the consequences of a certain treatment. Cancer detection and therapy are another prominent use of gene expression profiling.
- Cancer is a disease marked by uncontrolled cell multiplication and expansion. This arises as a result of mutations in the genes that control cell development. This suggests that mutated genes can be found in all malignant cells. To offer a more precise categorization of malignancies, gene expression profiling is used. The challenge of classifying gene expression data samples into various classes difficult. The number of genes in a typical gene expression dataset might range from a few thousand to tens of thousands.
- Learning classifiers are frequently used to solve this challenge, as they develop a population of condition action rules that drive the classification process. The eXtended Classifier System (XCS) has been effectively used in the biology and computer science sectors to classify big datasets. The version of XCS, COXCS, splits the whole search space into subdomains and uses the normal XCS algorithm in each of them. Because the classification issues on the subdomains may be handled concurrently, such a procedure is computationally costly yet readily parallelized.
- Cloud-CoXCS is a cloud-based CoXCS solution that uses Aneka to solve classification problems in parallel and assemble the results. The algorithm is guided by strategies, which specify how the outputs are combined and if the process is complete.


Comments
Post a Comment